How <PubNote> supports life sciences researchers using XML for OLSPub
Abstract
In May 2025 ZB MED in Germany announced OLSPub – the Open Life Science publication database – an open, reliable, and sustainable alternative to the US-based PubMed database of biomedical research publication metadata:
https://www.zbmed.de/en/research/current-projects/olspub
The <PubNote> GitHub repository then was created by Réalta Online Publishing Services Ltd. as a set of simple tools to assist OLSPub users of PubMed metadata XML files. These tools recognize that having to deal with raw XML markup can be discomfiting to those allergic to angle brackets:
https://github.com/RealtaOnline/PubNote?tab=readme-ov-file#readme
An important property of the rendering tools in this package is the off-the-shelf running implementation of sustainable single-source publishing practices. This sustainability concept was introduced at JATS-Con 2023 in contrast to traditional relatively unsustainable single-source publishing practices. <PubNote> demonstrates the success of these practices when OLSPub users trigger human-oriented visualizations of PubMed XML metadata in PDF, HTML, and DOCX syntax for their review in their favourite office tools.
This paper first introduces the new OLSPub resource and how it services the life sciences research community.
The <PubNote> suite of simple tools is described, illustrating the results available to users.
Sustainable single source XML publishing is reviewed, and the open-source tools supplied in the <PubNote> repository that implement sustainable single source publishing are enumerated and demonstrated.
Table of contents
OLSPub: a biomedical research metadata database for global access
The Open Life Science Publication database (OLSPub) infrastructure project was proposed by the German medical library ZB MED in May 2025 and is expected to be approved and funded before the end of 2025. The OLSPub project addresses the need for an open, multilingual, and transparent resource of biomedical research citations, supporting unfettered global access into the future:
-
Title: Strengthening the Biomedical Research Community by Building a Resilient and Sustainable Solution
-
https://www.zbmed.de/en/research/current-projects/olspub
The aim of the project is to strengthen the resilience and independence of the life sciences research infrastructure in Europe.
The database is initialized with the entire content of the long-established PubMed database of citations. This database is centrally controlled by the National Library of Medicine (NLM), a part of the National Institutes of Health (NIH), a part of the US government. Highlights of the evolution of PubMed over almost 150 years include:
-
1879-2004 – Index Medicus (in print form) of medical research information
-
1963 – computerization through MEDLARS (literature analysis and retrieval)
-
1968 – National Library of Medicine moved under National Institutes of Health
-
1971 – MEDLINE (MEDLARS online) metadata
-
1996 – experimental PubMed web-based interface to MEDLINE
-
1997 – official launch of free public access to all of MEDLINE via PubMed
-
2002 – core system switch to direct processing of XML-based metadata:
-
2025 – 39 million biomedical research citations (~2 million per year)
While there are some articles cited that are in other languages, PubMed is functionally Anglo-centric. OLSPub endeavours to accommodate researchers using other languages, both for input and output.
XML syntax for two semantic vocabularies for biomedical research metadata is used for getting content into and out of the PubMed database:
-
PubMed In – submission XML DTD (database ingress)
-
PubMed Out – distribution XML DTD (database egress)
The OLSPub project plan includes establishing a working group for the maintenance of the XML tagging guidelines for the use of these two vocabularies.
<PubNote>: a suite of XML assist tools for OLSPub users
The use of XML syntax is ideal for unambiguously labeling content going in to and out from a database of richly structured metadata information. However, as good as XML is for the computers, there are many human users with an allergy or aversion to the use of angle brackets. And they are not alone, as such is true in many XML projects.
The <PubNote> project, freely available on GitHub at https://github.com/realtaonline/PubNote?tab=readme-ov-file#readme (or https://bit.ly/ro-pubnote using a shortcut), is an open-source project in support of OLSPub and PubMed users to help them in the exposition of XML documents in office-friendly formats. The initial version was developed by Réalta Online Publishing Services Ltd. in Ireland, a provider of online publishing services for converting complete JATS XML documents to PDF, HTML, and DOCX formats for worldwide clients.
Applying lessons learned from publishing deeply-nested structured XML documents, the <PubNote> project takes as input either the PubMed In XML or the PubMed Out XML shallow metadata documents, and renders exposition outputs in PDF, HTML, DOCX, and indented XML. Moreover, the user can select a rendering that translates the XML element names into content heralds of different languages, initially English, German, and French:
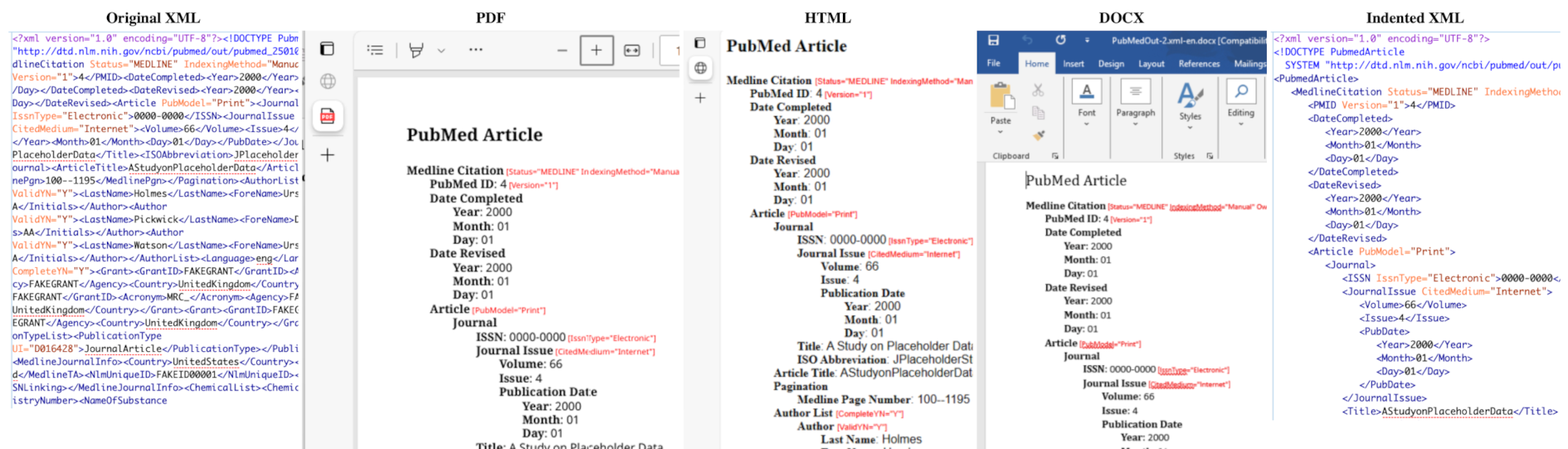
Anticipating that users needing help with the XML will need easy-to-use invocation of the tool, the four interfaces to <PubNote> are the DOS command line, the Shell command line, Windows drag-and-drop, and Mac drag-and-drop:
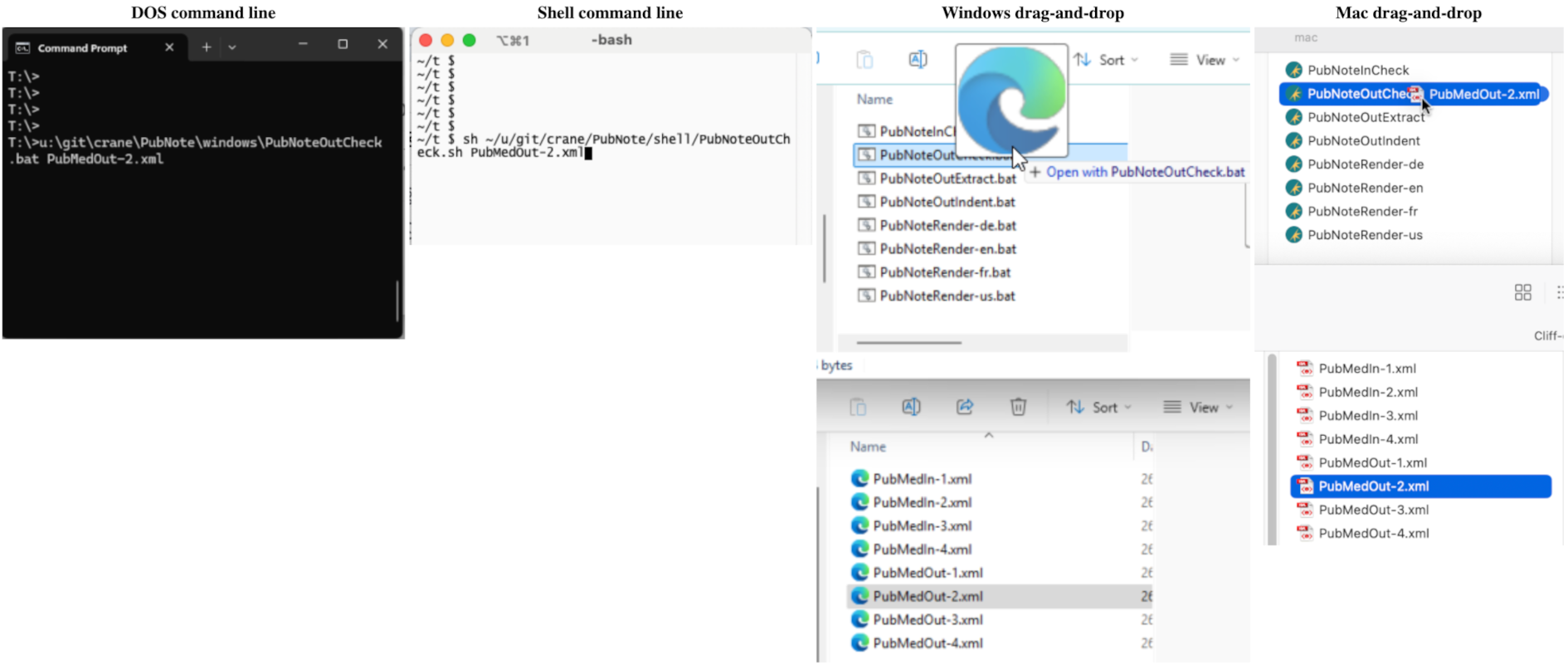
A single invocation produces all of the renderings of the selected language into a subdirectory named from the base filename of the input XML document, in the same directory as the input XML. Should there be any errors, a log file is placed in the same directory as the input XML. When there are no errors, no log file remains after the invocation completes.
In the plans for the future is a text-oriented output of the content of the XML to be used in conjunction with an as-yet-developed Invisible XML (iXML) script supporting round-tripping of the content back to XML. Some research is needed in this area to ensure a user experience supporting the editing of the XML without ever seeing an angle bracket.
Sustainable single-source publishing example
The concept of “sustainable” single-source publishing was introduced at JATS-Con 2023 in the https://www.ncbi.nlm.nih.gov/books/NBK591971/ presentation.
The promise of single-source publishing has been to reduce the effort in the creation process to create multiple renditions of a given concept. Consider the need in this project to produce PDF, HTML, and DOCX outputs.
Whereas three creative processes would originally be needed to produce three renditions, a single creation process that produces XML would be passed to a publishing process that produces the three renditions. Given that this project already has the input in XML as a single source, it would seem obvious that traditional single-source publishing would be deployed to produce the three outputs.
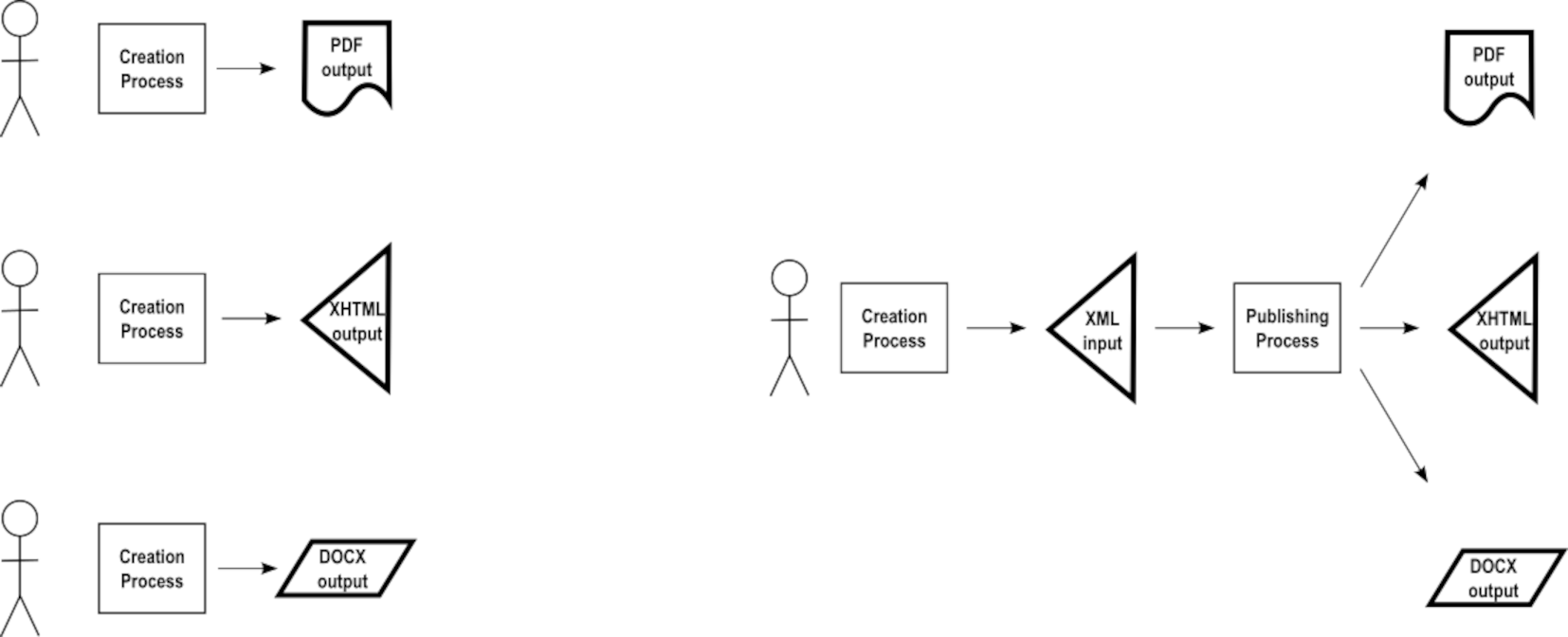
However, that one “Publishing Process” box often depicted for single-source publishing in fact breaks down to separate publishing tasks for each of the outputs, as shown here. A transformation for each of the outputs reads the XML source to produce the expression needed to generate the output format desired.
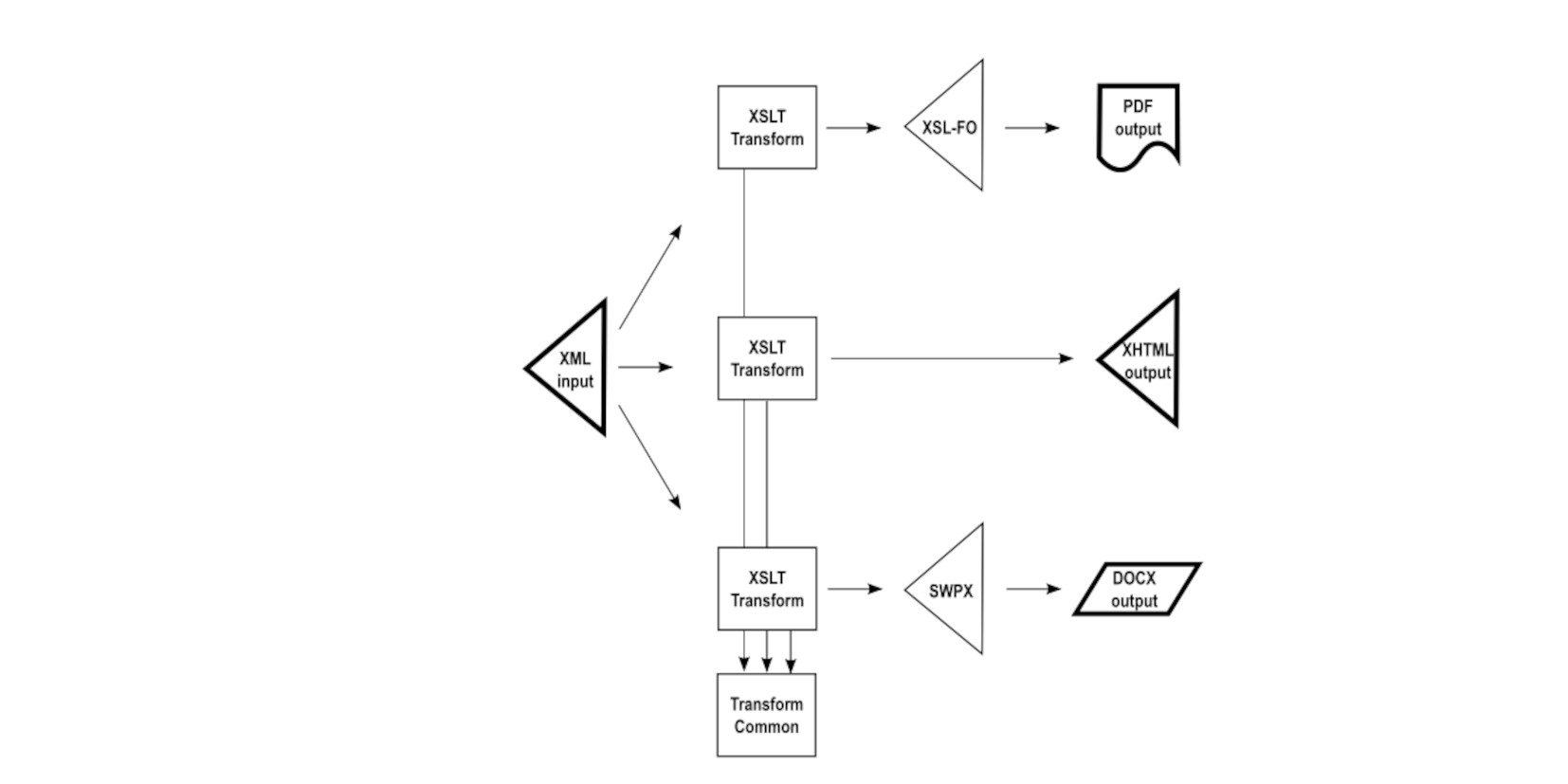
Forward-looking implementations often distill out from each of the transformations common bits of shared logic. Typically these common bits are abstracted strings or logic not producing the output directly. Rather, they are invoked by each of the independent transformations in a particular context needed at output generation time. This approach promotes consistent results between the output formats, but does not guarantee fidelity because the common bits are invoked in different contexts for the different outputs. These may not be perfectly consistent.
This lack of assurance of fidelity between renditions is rooted in the need for each of the transformations to be a semantic interpretation of the source XML. Should the semantic interpretation vary over time, it is necessary to update, in this example, three separate transformations to reflect the required logic. When something can go wrong, it may go wrong, and it may happen that each of the transformations implements different interpretations of the semantics rather than the same.
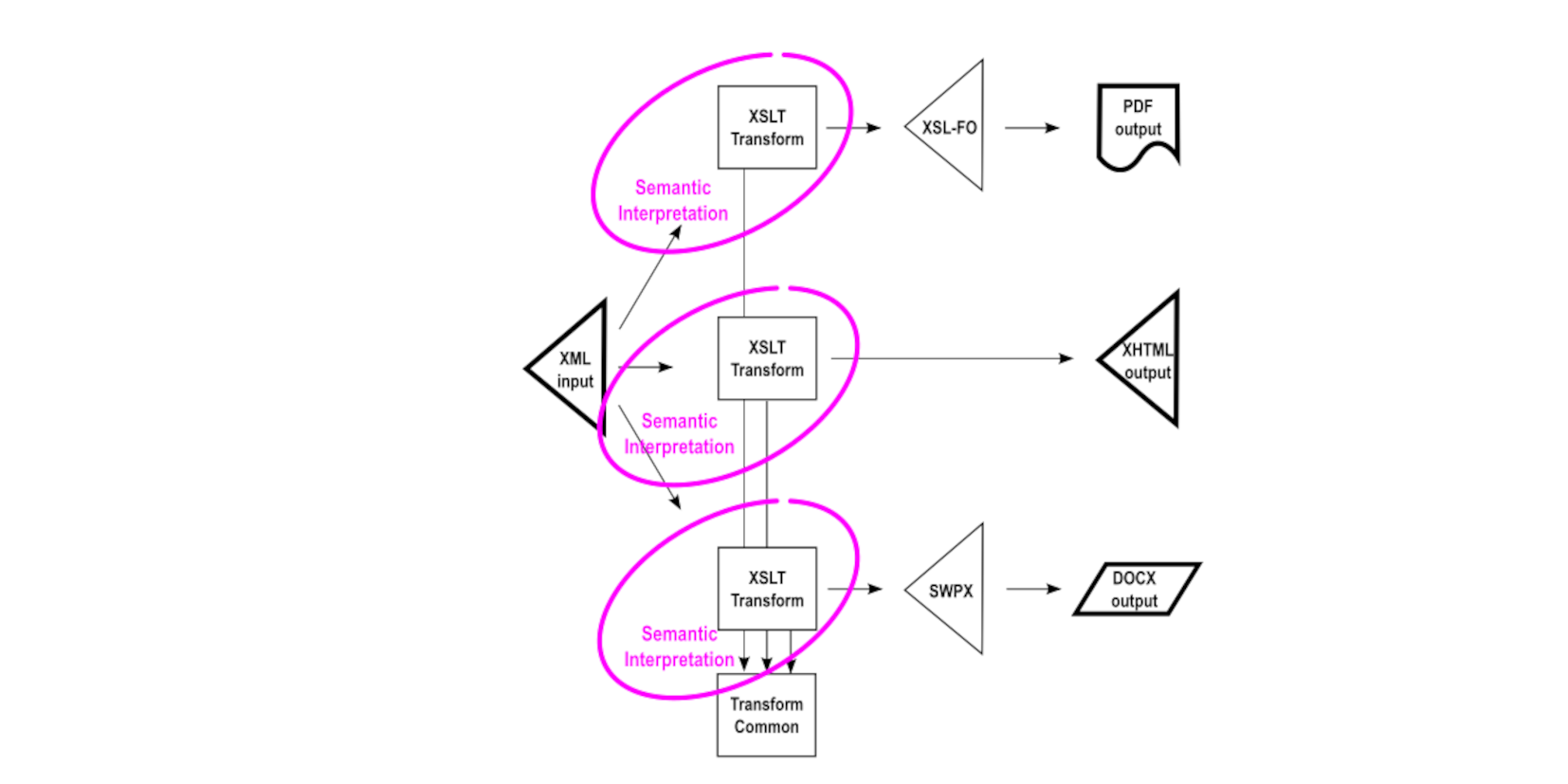
This issue is illustrated in more detail as follows, showing how each of the semantic interpretation transformations produces different output markup vocabularies. The <PublisherName> element is interpreted as a <block> in XSL-FO, as a <div> in XHTML, and as a <wp:p> in SWPX (the Wordinator Simple Word Processing XML vocabulary used to produce DOCX).
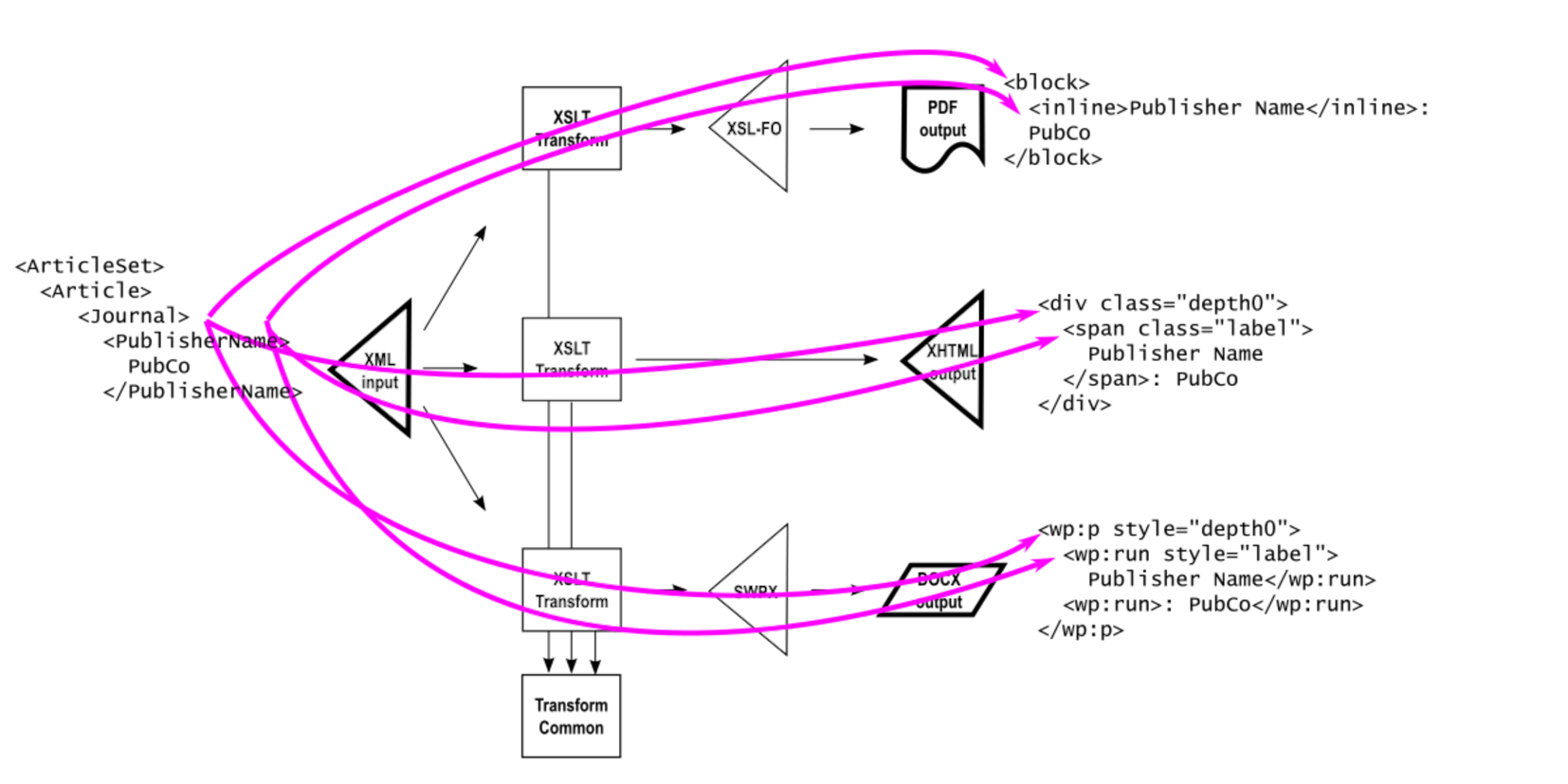
Note how in each of the three transforms, the stylesheet writer has had to interpret the semantic need for the prompt to be “Publisher Name” (with a space). If by a slip of the fingers any one of these is misspelled, there will be infidelities in the result.
And so it is when requirements in semantic interpretation change. It is incumbent on the programmer to implement the semantic logic three times in parallel in an attempt to preserve fidelity. This can be unsustainable over time.
A sustainable approach to single-source publishing is to get back to the original principle of doing something only once. In such a data flow, the output from one transformation is used as the input to the following transformation.
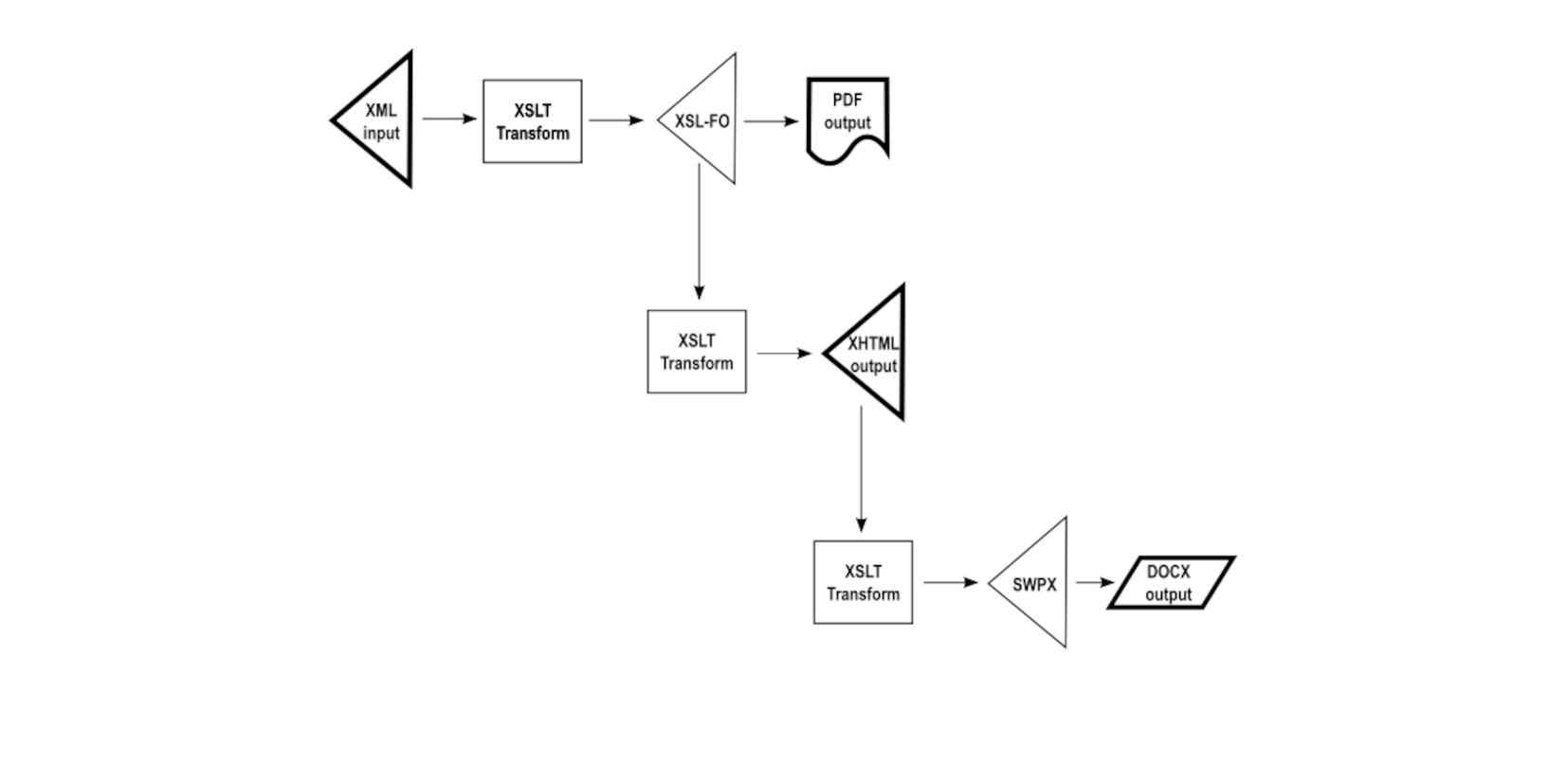
This may not be obvious until one realizes that it is only the initial transformation that is obliged to implement the semantic interpretation of the source XML. Subsequent transformations can leverage the output of the initial transformation in mechanical fashion, without any semantic knowledge of the original input source. This promotes, and practically ensures, content fidelity between the set of output renditions, reducing any opportunities for inconsistent semantic interpretation.
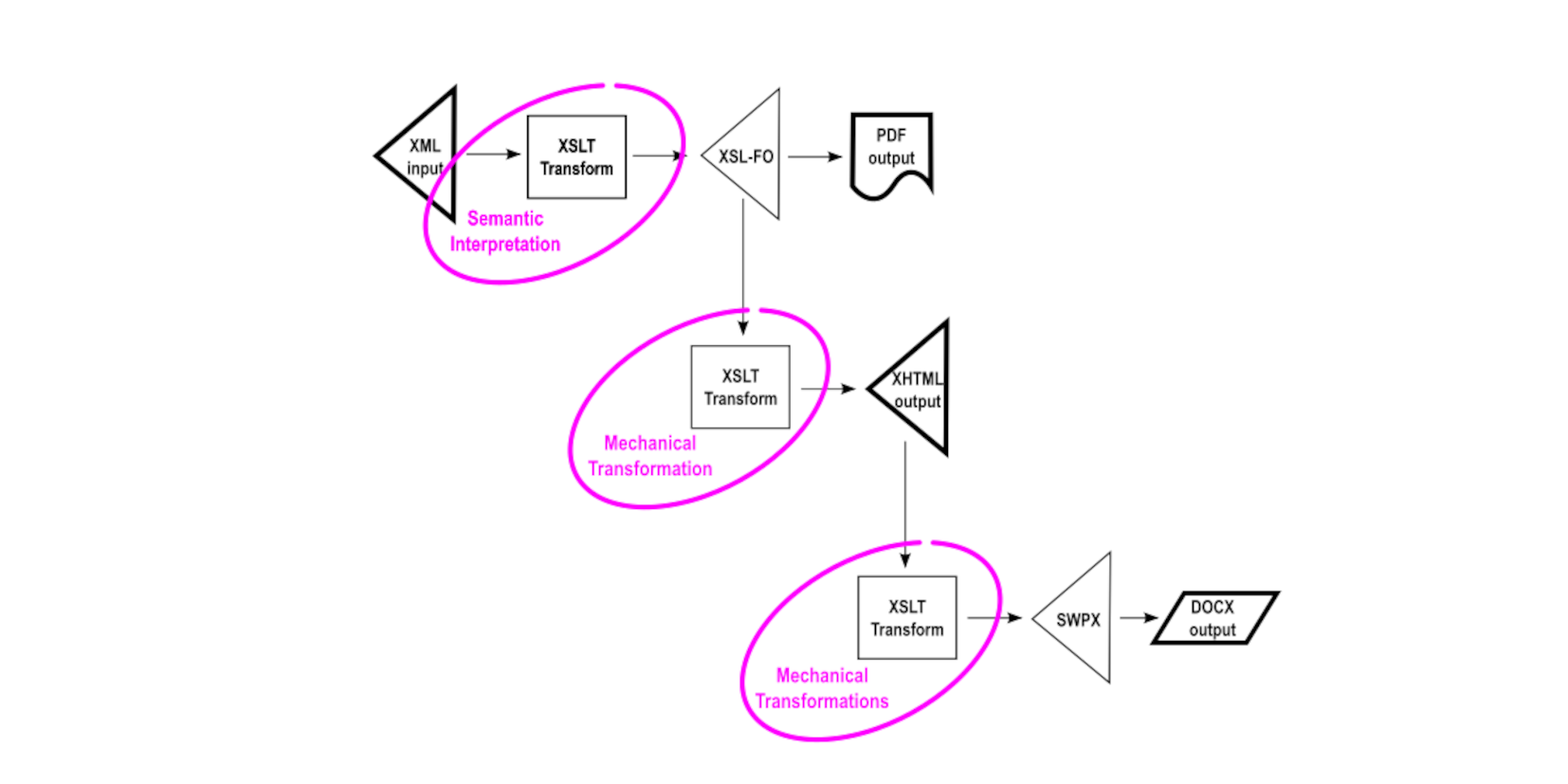
To illustrate the distinction between semantic and mechanical interpretation, consider how the initial semantic interpretation of <PublisherName> into a block surrounding the string “Publisher Name” includes at the same time some information intended for downstream use.

A conforming XSL-FO processor is silently tolerant of non-XSL-FO constructs in foreign namespaces found in the XML instance. Such are ignored. In the <PubNote> project, attributes in a <PubNote> namespace are injected into the output XSL-FO at semantic interpretation time. In this case, the attribute p:h="div*depth0" is added as part of the block, and the attribute p:h="span*label" is added to the inline construct surrounding the data herald being output.
This attribute encoding indicates to downstream processes the XHTML element name before the asterisk, and the CSS style name after the asterisk. As a stylesheet writer doing the semantic interpretation of the input XML, the downstream accommodations are considered at the time the semantics of the input XML are being interpreted.
After this point, transformations are mechanical: the XHTML+CSS is synthesized from the contents of the p:h= attributes, and, in this project, it is enough to name the Word styles the same as the CSS styles. SWPX is likewise a very flat representation of information as HTML, rather than a deep hierarchical structure, and so it ends up being very straightforward to mimic the output flat structure for the final stage.
Note how this illustrates how the fidelity between the three outputs not only is promoted, but practically guaranteed, in regards to the English data herald “Publisher Name”. The semantic interpretation has determined the need for “Publisher Name” and put that into the XSL-FO output. The XHTML simply copies the text input of the XSL-FO, and so typographical errors on behalf of the stylesheet writer are possible. Likewise, the SWPX simply copies the text input of the XHTML, thereby guaranteeing fidelity in the data herald across all three outputs. Of course, if there is a typo in the original semantic interpretation, that will get propagated to all of the subsequent outputs.
Moreover, but not illustrated here, the mapping of the element name “PublisherName” to the data herald “Publisher Name” is done at semantic interpretation time through a lookup table, rather than being hard-coded as illustrated above. This approach provides the opportunity to offer the presentation of PubMed content using data heralds of different languages. In the first release, three <PubNote> semantic interpretation stylesheets invoke common code and distinct lookup tables for end users to inspect the content of XML documents using data heralds in English, German, and French.
For more detail and another example of this sustainable single source publishing, see https://www.ncbi.nlm.nih.gov/books/NBK591971/ for a complete paper published in the proceedings for JATS-Con 2023.
The <PubNote> data flows and tools
<PubNote> attempts to offer the users of PubMed submission and distribution XML instances many features in being able to consume the information found necessarily encoded in streams of angle brackets. This data flow diagram illustrates how information going into and out of either OLSPub or PubMed can be massaged using the <PubNote> suite of utilities:
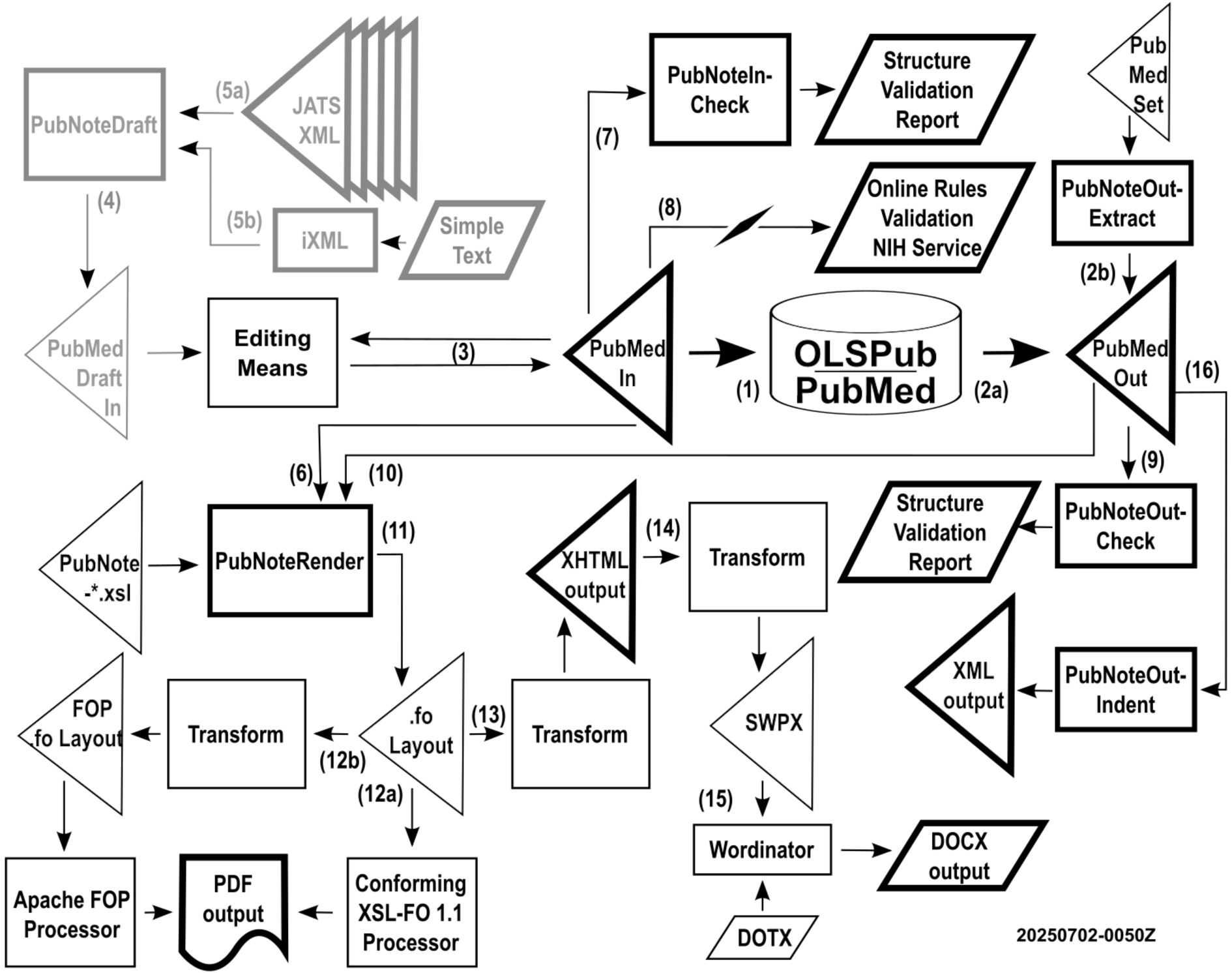
A valid PubMedIn XML submission document is forwarded for inclusion in the OLSPub and/or PubMed database
A valid PubMedOut XML distribution document is obtained for processing
2a. exported directly from the OLSPub and/or PubMed database according to some criteria, or
2b. extracted (with or without added indentation) using the PubNoteOutExtract process against one or more valid PubMedOut XML distribution documents, such as from an ~200MB XML single article set document of 30,000 article documents unpacked from a ~20MB ZIP file from the PubMed FTP site
Presumably, there is some editing process for the user's PubMedIn submission document
Optionally, the user uses PubNoteDraft to create a PubMedDraftIn document as a starting point (Phase 2)
Inputs used to create draft are expected to include (among others):
5a. – a distillation of multiple JATS XML documents into a draft, or
5b. – a simple text file with no angle brackets converted to XML using iXML (Invisible XML)
For human review, PubNoteRender formats the information found in a PubMedIn document
Optionally, PubNoteInCheck can validate a PubMedIn document against the PubMed.dtd
Optionally, a PubMedIn document can be checked using the NIH online business rules validator
Optionally, PubNoteOutCheck can validate a PubMedOut document against the pubmed_250101.dtd
For human review, PubNoteRender formats the information found in a PubMedOut document
XSL-FO is created with all of the content to be presented downstream in PDF, HTML, and DOCX
XSL-FO is transformed into PDF for print purposes
12a. – a conforming XSL-FO 1.1 processor directly produces the output (not part of this repository)
12b. – a non-conforming XSL-FO 1.1 process requires a transformation to make a suitable subset XSL-FO
XSL-FO is transformed into XHTML for browser display
XHTML is transformed into SWPX (Simple Word Processing XML) for word processing purposes
The Wordinator tool combines SWPX and a Microsoft DOTX template to produce a DOCX file
Optionally, PubNoteOutIndent processes a PubMedOut document into an indented XML output
The tools used in implementing these data flows are found in the utilities/ project subdirectory.
XSLT stylesheets are executed using the Saxon 12HE (Home Edition) processor. The stylesheets are documented using Crane Softwrights Ltd.’s XSLStyle documentation methodology and stylesheets.
The Jing processor is used for XML instance validation against a Relax-NG (RNG) version of the two PubMed DTDs. The RNG was created using Trang (not included in the project).
The Apache FOP XSL-FO engine is included, but it is not conforming with respect to the presence of elements and attributes in foreign namespaces. Accordingly, there is a filtering step to remove the annotations before handing the XSL-FO instance to FOP for processing to create the output PDF files.
For conversion of SWPX to DOCX, the Wordinator distribution by Eliot Kimber is included, plus a separate directory in which is found a “fat” version of the JAR file complete with all dependencies in order to run standalone.
Conclusion
The biomedical research community needs the reliable and full-featured OLSPub database with which it can move forward with its invaluable progress towards solving our world’s medical challenges. And it can do so with the fidelity of PubMed content without worry about what changes there may be in the future regarding access to the original US-based resource.
XML is an unambiguous syntax with which one labels the content of a stream of information such that computer systems can identify portions of that content by the labels. In turn, using those labels to identify commonly-understood semantics behind the labeled content, computer systems can effectively interpret the stream of content for its intended purposes. This is ideal for the description of database information, and so XML is used for both ingress and egress of bibliographic citation information.
The successful deployment of OLSPub is supported with suites of XML tools that help this community, who may not be familiar with or experienced with the vagaries of rigid XML syntax.
The <PubNote> project is only one such suite of XML tools. It provides multi-lingual exposition of instances of PubMed submission XML and PubMed distribution XML documents. The project is open to participation to anyone interested in contributing to the success of OLSPub by improving on the features.
